Singular value decomposition.
A powerful mathematical tool that arises from linear algebra is the “dimension reduction” method using the so-called singular value decomposition (SVD). This example is written in a tutorial style.
Matlab/Octave
In this set of exercises you will be using either Matlab or its libre
counterpart Octave. It is advised to open a script file
(File -> open -> create a script), and code there. If
you run the code by pressing the “run” button (play icon). You can
inspect the declared variables in the command-line section. This
document will involve some coding, but I hope you will be able to follow
the code snippets in this document. You may copy/paste and extend
them.
Data in a square matrix
We can store all kinds of information in columns and concatenate columns to form a data matrix. We start by considering an artificial example: A researcher in biomedical engineering analyses data from patients. It so happens to be that he/she is interested in quantities regarding these patients. We illustrate the first patient’s column vector .
For 3 patients, we can place all the data in a matrix , The rows imply the meaning and units.
This matrix maybe diagonalizable (why?). We can use
Octave/Matlab to quickly find its eigen values and eigen
vectors:
% Initialize matrix
A = [[45, 40, 50],
[180, 172, 173],
[2.2 , 2.3 , 2.1]];
% Compute eigen vectors and eigen values
[V, l] = eig(A);1)
Using the code below, verify that V and l are
the “eigen decomposition” of
.
Hint: use inv(A) to find the inverse of a matrix
A.
% Compute the reconstructed matrix `Ar` using the eigen decomposition
Ar = V*l*inv(V); 2) Using the code below, verify that the eigen vectors are normalized
% Lets have a look at the first eigen vector
eig1 = A(:,1);
% We can square the components and sum them to get the squared length
% Mind the dot in the element-wise square operator `.^2`.
leig1 = sum(eig1.^2); From inspection of the matrix l, the matrix
has three distinct (real!) eigen values, from large to small:
It appears that
is much larger than the others. Imagine if we would set all values in
the diagonal matrix (l) to zero, except for
.
We can call this a “low rank approximation” of the original matrix.
% Row-rank approximate diagonal matrix
l_lr = zeros(3,3);
l_lr(1,1) = l(1,1);Instead of using the original decomposition (), we can attempt a low-rank approximation of the data set.
% Low rank approximation of A
A_lr = ...%fill inUsing the rules of matrix multiplication, we can see that the second
and third column of V do not contribute to this product.
Similarly, the second and third row of
play no role in this approximation. As such, we can approximate the
matrix
,
using fewer data. The approximation requires 3 + 1 + 3 = 7 non-zero data
entries, instead of the original 9 (3 times 3). This is a “lossy” data
compression method, as we are not able to reconstruct the original data
matrix exactly.
3) Rewrite the low-rank approximation as a column vector - row vector multiplication.
4) Have a look at the component values in the first eigen vector (associated with ). What do these numbers tell you about an “Eigen person”?
Notice that we could make a better approximation if we would include the second largest eigen value, and its associated column and row in and , respectively.
Data compression in a rectangular matrix
Grey-scale image data consist of a grey-scale value for each pixel in the image. We can import an image and convert it to a grey-scale matrix using the following code in Octave/Matlab. (replace the link with a non-square (moderately sized) image/photo that you like).
% Load an image via a path or the internet
[Image,b] = imread ("https://assets.w3.tue.nl/w/fileadmin/_processed_/\
3/f/csm_Gemini_zuidzijde_396-fr2-a0002_web_0b845bfc46.jpg");
% The color Image is now represented as a `m x n x 3` matrix.
% We should average over the 3rd dimension (RGB-values) to obtain a grey-scale image
grey = mean(Image, 3);
% Inspect the grey-scale image using,
size (grey)
imshow (grey, b);
Since the image it not square, we cannot readily apply the eigen-decomposition method above. However, and are square, and these will appear to encode (some) of the data of our grey-scale image.
We will strive to find the matrices that decompose a matrix , according to.
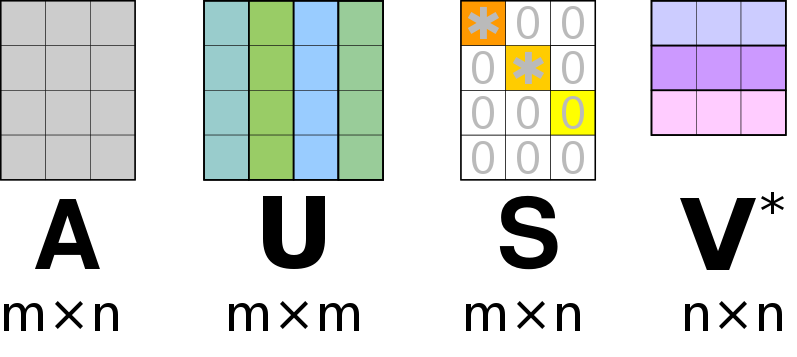
Similar to the eigen-value decomposition will be a diagonal matrix. For an matrix, the matrix will be of size , the diagonal matrix will be of size and will be a matrix. For an SVD () decomposition, we dictate that the columns of and must be normalized, this ensures that the diagonal entries of are the “weights” associated with the corresponding columns of and . Furthermore, the ansatz is that the columns in and are orthogonal. An auxiliary theorem (not part of this course) states that for an orthonormal matrix (), its inverse is equal to its transpose;
Lets derive an expression that relates , and .
And analogous,
5) Proof that
We can see that the columns of and are the eigen vectors of and , respectively, with the eigen values the associated entries of the diagonal square matrices and , respectively.
Our ansatz that the columns of and are orthogonal is hereby closed. This is rooted in an auxiliary theorem (not part of this course) that states that the distinct eigen vectors of a symmetric matrix () are orthogonal.
6) Show that and are symmetric matrices.
A failing attempt
The next computer coding is related to finding the eigen values and
eigen vectors for
and
.
We can use Octave’s eig function.
% Compute the two square matrices
AAT = grey*transpose(grey);
ATA = transpose(grey)*grey;
[U, STS] = eig(AAT)
[V, SST] = eig(ATA) 7) Inspect
the sizes of the matrices U, V,
STS and SST
8) Inspect the eigenvalues using,
plot (abs(diag(STS)))
hold on
plot (diag(SST))
set(gca, 'YScale', 'log')
hold off; We should ensure that the eigen values appear in descending order and also reorder the eigen vectors accordingly.
% Sort eigen values and vectors
[sst, idx] = sort(diag(SST), "descend");
Us = U(:,idx);
[sts, idx] = sort(diag(STS), "descend");
Vs = V(:,idx);9) Compare the eigen
values sst and sts
Now we should compute .
10)
Show that the diagonal entries of
(i.e. the singular values) are the square roots of the eigen values
stores in sts and/or sst.
We can now compute ,
% Compute the singular-value diagonal m x n matrix (S)
s = sqrt(sst);
sg = size(grey);
S = diag(s, sg(1), sg(2));We are now ready to test our singular value decomposition
11) Inspect the sizes of U, S, V, the singular values, and, using the code below, verify the decomposition.
gr = U*S*transpose(V);
imshow (gr, []);
A successful decomposition
Indeed, when we discovered that both and , we have wrongly assumed that these equations ensure that the original decomposition is automatically satisfied. This would have worked if the singular value decomposition is unique. But there may exist many matrices () that are smilar to in the sense that etc.
Instead, we should consider the more intimate connection between , and the original matrix () given by,
12) Derive this expression.
Given that
is a diagonal matrix, we see that the linear transformation of the the
matrix
,
results in a matrix with scaled columns of
.
At this point we can choose to first compute an
and
using the eig() function (as we already did), and then
solve for the intimate relation above to obtain the associated
.
Alternatively, we could compute
and
using eig() and solve for the associated
.
Both are fine, but it should be clear that if we have
,
and
,
we can readily compute the product
,
and since we know the diagonal matrix
,
the columns of
are the columns of
scaled by the diagonal entries of
.
Since
is a diagonal matrix, apply the scaling operation to each column of
in one operation in MATLAB/Octave.
% Compute Uv that will decompose A, using V and S
Uv = A*V/S;Noting that this Matalb/Octave specific syntax should not be confused with proper matrix algebra (You can not divide by a matrix!).
Now we can test the decomposition
13) Inspect the sizes of U, S, V, the singular values, and, using the code below, verify the decomposition.
gr = Uv*S*transpose(V);
imshow (gr, []);
Data compression.
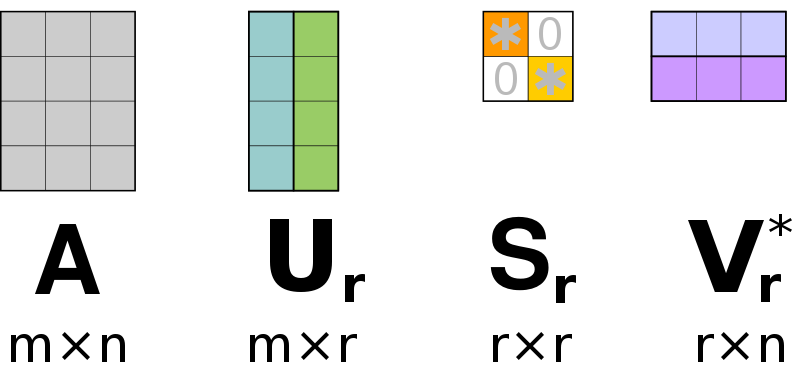
In a final step, we select only the most prominent columns of () and () that are associated with the largest singular values in ().
14) What are the sizes of the matrices , , and ?
We can use some Matlab/Octave coding to shrink the matrices.
% set a low-rank value
r = 16;
% First r columns of U and V form Ur and Vr
Ur = U(:, 1:r)
Vr = V(:, 1:r)
% We should only keep the associated rows and columns of $S$ as well
% Such that we only keep the upper left r x r entries of S.
Sr = S(1:r, 1:r)
% Approximate the image using only $j$ singular vectors in both Ur and Vr
gs = Ur*Sr*Vr';
imshow(gs, []);15) Try a few different values of and enjoy your low-rank approximations

